Peter Beerli

Institutional home page
http://people.sc.fsu.edu/~beerli
Arrival and Departure
May 22 - June 2, 2025
Lecture Materials
Note: The presentations may change up until the moment the lecture or lab begins. I will try to post the modified the PDF before the lecture but will post the actual talk-PDF after each talk.
Coalescence lecture (Monday, May 26 2025): I will introduce the coalescent and its use for population genetics inference. The first section of the lecture will discuss basics; in the second part I will discuss extensions of the basic coalescent. I will focus on neutral population processes, such as population structure, population size changes through time, population divergence, and problems and complications with inferences using the coalescence.
- May 26 (14:00-15:30): Introduction to the coalescent Coalescence2024-talk1 (talk given earlier: Coalescence2018-talk)
- May 26 (15:30-17:00): extending the basic coalescent Coalescence2024-talk2
- May 27 (9:00-12:00) MIGRATE tutorial and evolutionary model selection beyond mutation models, specific tutorial instructions Tutorial-Intro Tutorial
Teaching Tools
- Bugs_in_box is a python3 program that visualizes coalescence of bugs (well they are ladybugs and beetles) in a box. They crawl like crazy until they hit another bug, devour it and move on. There is a help menu (press H) that displays all options. This application has a few shortcomings: (1) it does not adjust well for different screen size (for example presentation beamers make the bugs huge); (2) No way to save or print directly out of the app, but Dave Swofford contributed a fun spin when there are two bugs left (see help menu in python bugsinbox.py).
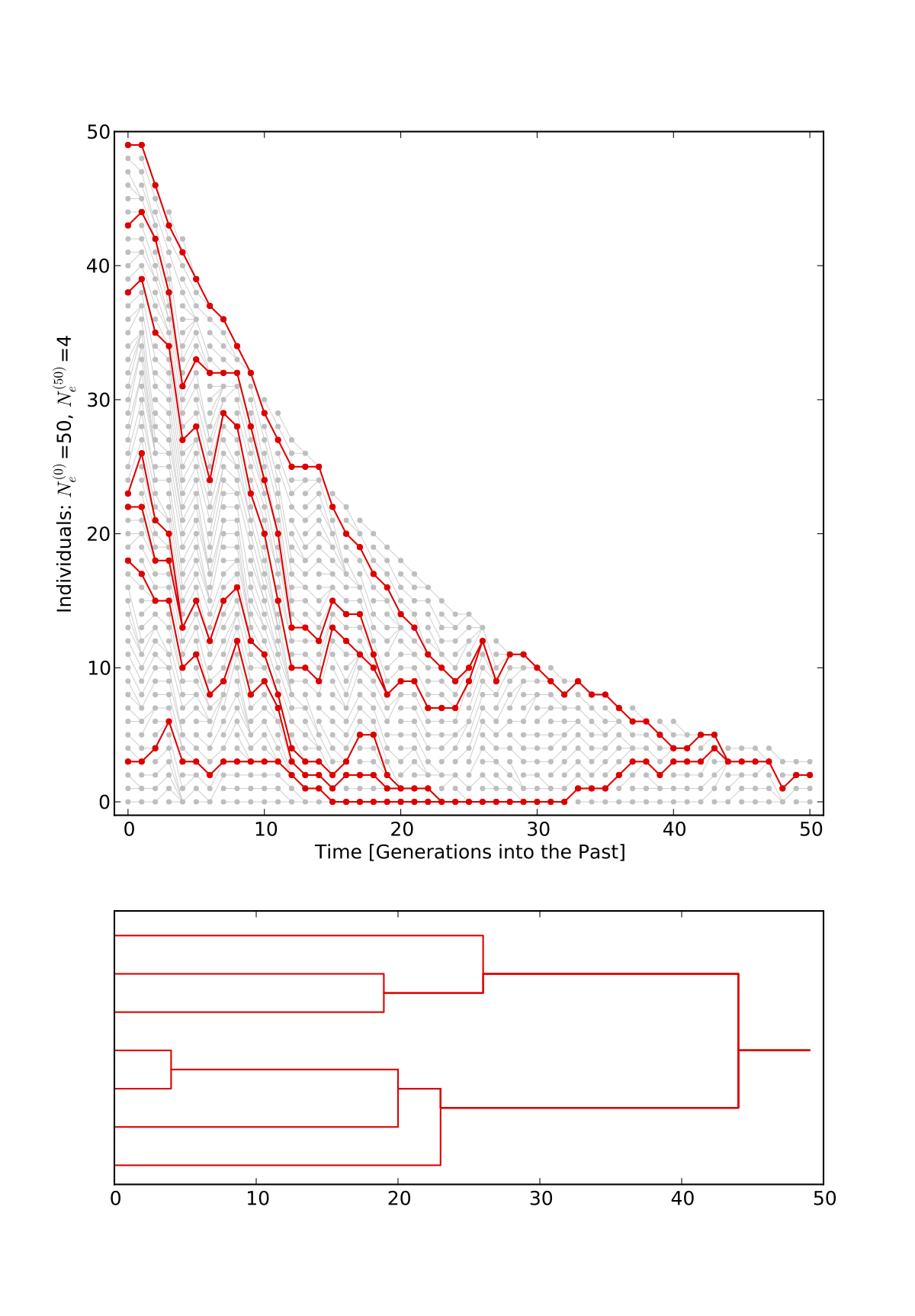
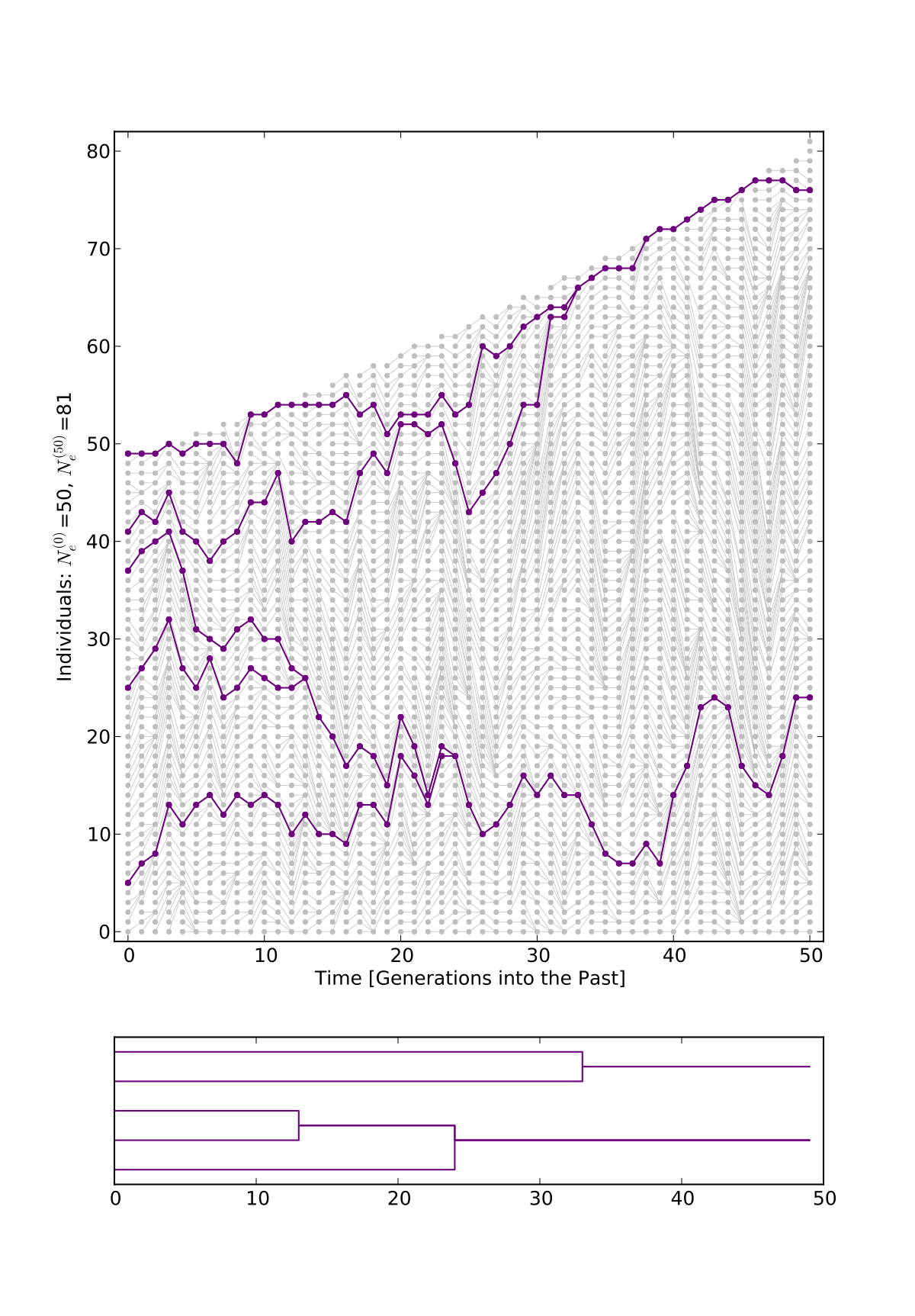
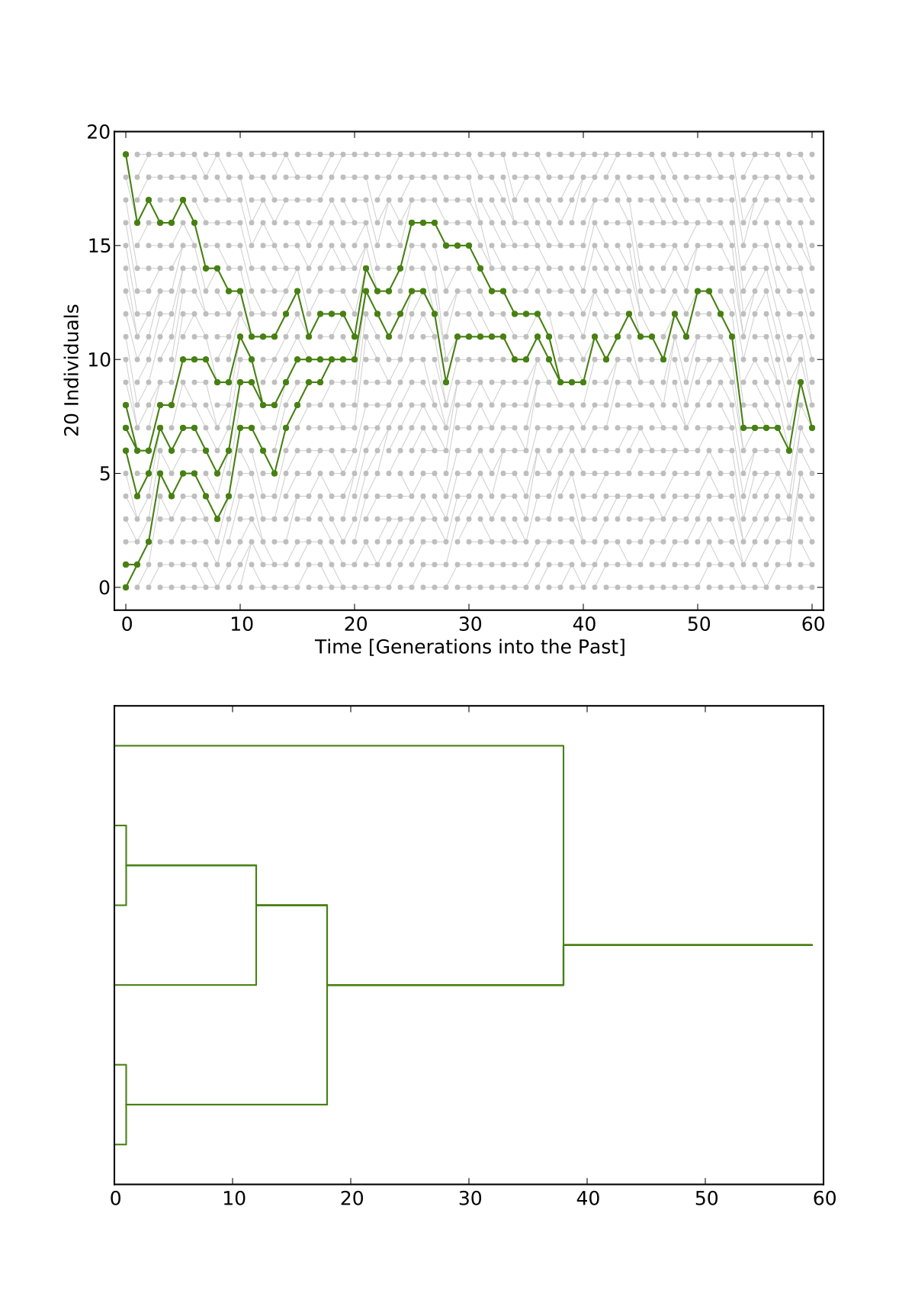
- PopVizard is a python program that allows to plot pictures of population genealogies like the ones on this page.
Demo and Tutorials
- Population-model selection using MIGRATE on May 27 2025. The tutorial guides you on estimating the most likely path of the Zika virus and discusses estimation of divergence times, gene flow, and model selection with MIGRATE 5.0.6. An older gene flow tutorial using the older version of migrate 3.x: PDF).
- If students are interested I am also happy to talk about LAMARC and give a Lamarc tutorial: Recombination estimation with LAMARC
Bayes factors and effect of parallel runs of MIGRATE
-
Lecture on Bayes factor that may help to understand the tasks in the MIGRATE lab. MIGRATE can run on many different operating systems and hardware, most efficiently it can run on computers that have more than one core (almost all these days) using parallelization:
-
Lecture on parallel installation of MIGRATE and Slice sampling.



